EXAONE Path 2.5
[Github] [Hugging Face] [Paper] [Cite]
Introduction
EXAONE Path 2.5 is a biologically informed multimodal framework that enriches histopathology representations by aligning whole-slide images with genomic, epigenetic, and transcriptomic data. By enabling all-pairwise cross-modal alignment across multiple layers of tumor biology, the model captures coherent genotype-to-phenotype relationships within a unified embedding space. This domain-informed design improves resource efficiency, enabling the model to achieve competitive performance across diverse tasks while using substantially fewer training samples and parameters than existing approaches.
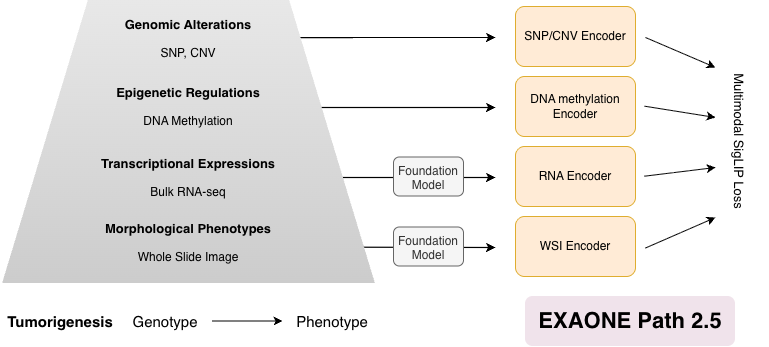
Figure 1. Overall scheme of EXAONE Path 2.5.
Quickstart
Load EXAONE Path 2.5 and extract features.
1. Hardware Requirements
- NVIDIA GPU with 12GB+ VRAM
- NVIDIA driver version >= 525.60.13 required
Note: This implementation requires NVIDIA GPU and drivers. The provided environment setup specifically uses CUDA-enabled PyTorch, making NVIDIA GPU mandatory for running the model.
2. Environment Setup
First, install Micromamba if you haven't already. You can find installation instructions here. Then create and activate the environment using the provided configuration:
git clone https://github.com/LG-AI-EXAONE/EXAONE-Path-2.5.git
cd EXAONE-Path-2.5
micromamba create -n exaonepath python=3.12
micromamba activate exaonepath
pip install -r requirements.txt
3. Inference Workflow Overview
EXAONE Path 2.5 inference follows a two-stage pipeline. (1) Patch-level feature extraction: extract pretrained patch embeddings from either image patches or full WSIs. (2) Slide-level feature extraction: aggregate patch embeddings into slide representations aligned with genomics data. Sections 3.1 and 3.2 describe these steps in detail.
3.1. Patch Feature Extraction
You can extract the pretrained patch features (without multimodal alignment) in two ways.
- 3.1.1 (Tensor output): for rapid prototyping or custom pipelines
- 3.1.2 (HDF5 file output): for full WSI processing, visualization, and downstream slide encoding
3.1.1. Tensor output
Assuming you have an image, you can run the following code snippet to extract pretrained patch features.
import torch
from PIL import Image
from torchvision import transforms
from transformers import AutoModel
repo_id = "LGAI-EXAONE/EXAONE-Path-2.5"
device = "cuda"
# Input
png_path = "path/to/your/sample_patch.png"
# Load patch encoder
patch_encoder_model = AutoModel.from_pretrained(
repo_id,
subfolder="patch-encoder",
trust_remote_code=True,
).to(device).eval()
# Image preprocessing (must match patch encoder training)
transform = transforms.Compose([
transforms.Resize(256),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
])
img = Image.open(png_path).convert("RGB")
image_tensor = transform(img).unsqueeze(0).to(device) # [1, 3, 224, 224]
with torch.no_grad():
patch_encoder_embedding = patch_encoder_model(image_tensor) # [B=1, C]
Outputs
patch_encoder_embedding: a tensor of shape[B=1, C]whereBis the batch size, andCis the embedding dimension- This tensor can be passed directly to the slide encoder in Section 3.2.
3.1.2. Full WSI patch-feature pipeline (HDF5 Output)
The step is further broken into smaller steps.
(1) Generate patch coordinates (and contour indices) with Python function API patchfy that you can import and call directly to:
- segment tissue regions
- extract patch coordinates
- (optionally) write a HDF5 file with
coords+contour_index
from exaonepath.patches import patchfy
wsi_path = "path/to/your/slide.svs" # .svs/.tif/.tiff/.ndpi/.mrxs/...
out_dir = "path/to/output_dir"
h5_path, coords, contour_idx = patchfy(
wsi=wsi_path,
out=out_dir,
patch_size=256,
step_size=256,
patch_level=0,
save_h5=True,
save_mask=True,
auto_skip=True,
)
Outputs
- If
save_h5=True, the patches are saved to:<out_dir>/patches/<slide_id>.h5
- If
save_mask=True, a segmentation visualization is saved to:<out_dir>/masks/<slide_id>.jpg
- If the slide is skipped due to the segmentation safety cap, a reason is written to:
<out_dir>/skipped/<slide_id>.txt
Note: the effective patch_size/step_size written to the HDF5 may be MPP-normalized internally (see the patchfy docstring for details).
The returned arrays are:
coords:N x 2int array of patch coordinates(x, y)in level-0 pixel space.contour_idx: int array of lengthNholding the tissue contour index of each patch.
Useful parameters
seg_downsample(float): extra downsampling factor for segmentation only (speed vs accuracy).max_seg_pixels(float): skip very large slides at the chosen segmentation level (set<=0to disable).- Advanced segmentation/filtering knobs: see the
patchfydocstring inexaonepath/patches/patchify.py. (e.g.,sthresh,mthresh,close,use_otsu,a_t,a_h,max_n_holes,line_thickness).
(2) Extract patch features from the WSI using those coordinates:
python -m exaonepath.feature_extraction.extract_single_slide_feature \
--slide_path "path/to/your/slide.svs" \
--coords_h5_path "<out_dir>/patches/<slide_id>.h5" \
--out_h5_path "<out_dir>/patches/<slide_id>_features.h5" \
--batch_size_per_gpu 32
Notes
coords_h5_pathmust be the H5 produced bypatchfy(save_h5=True). Future slide encoder requirescoords, ideally withcontour_index.- The output file (
out_h5_path) will contain:features[N, C],coords[N, 2],contour_index[N].
3.2. Slide Feature Extraction
Patch features, coordinates, (contour index) must be available. Use the below code snippet if patch feature extraction was conducted with 3.1.2. patch_features_h5_path should be identical as out_h5_path from the previous step.
import h5py
import torch
from transformers import AutoModel
device = "cuda"
# Load slide encoder (HF)
repo_id = "LGAI-EXAONE/EXAONE-Path-2.5"
slide_encoder = AutoModel.from_pretrained(
repo_id,
subfolder="slide-encoder",
trust_remote_code=True,
).to(device).eval()
# Load patch-level features exported as an HDF5 file
# Expected keys: features [N, C], coords [N, 2], contour_index [N]
patch_features_h5_path = "<out_dir>/patches/<slide_id>_features.h5"
with h5py.File(patch_features_h5_path, "r") as f:
patch_features = torch.from_numpy(f["features"][:]).float() # [N, C]
patch_coords = torch.from_numpy(f["coords"][:]).long() # [N, 2]
patch_contour_index = torch.from_numpy(f["contour_index"][:]).long() # [N]
patch_features = patch_features.unsqueeze(0).to(device) # [B=1, N, C]
patch_coords = patch_coords.unsqueeze(0).to(device) # [B=1, N, 2]
patch_contour_index = patch_contour_index.unsqueeze(0).to(device) # [B=1, N]
# All patches are valid (if you use padding later, set False for padded tokens)
patch_mask = torch.ones(
(patch_features.shape[0], patch_features.shape[1]),
dtype=torch.bool,
device=device,
)
with torch.no_grad():
outputs = slide_encoder(
patch_features=patch_features,
patch_mask=patch_mask,
patch_coords=patch_coords,
patch_contour_index=patch_contour_index,
)
patch_embedding = outputs["patch_embedding"]
slide_embedding = outputs["slide_embedding"]
If you padded patches and want to keep only valid patch embeddings (ragged), use the mask to index.
(This returns a Python list because N_valid can differ per slide.)
valid_patch_embeddings = [
patch_embedding[b, patch_mask[b]] for b in range(patch_embedding.size(0))
]
Output
patch_embedding:[B, N, C]updated patch-level embeddings after multi-modal alignmentslide_embedding:[B, C]slide-level embeddings to be used for downstream tasks
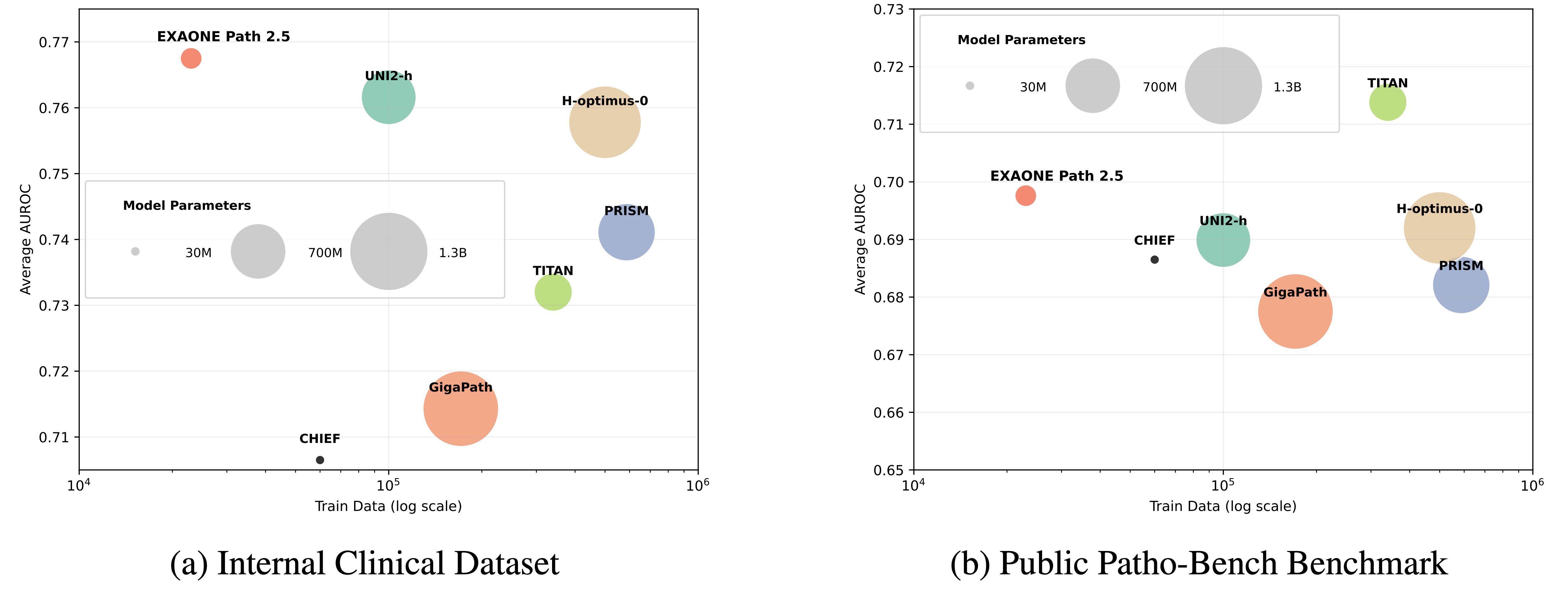
Model Performance Comparison
Experiments on internal multi-institutional clinical datasets and the 80-task Patho-Bench benchmark show that EXAONE Path 2.5 matches or surpasses state-of-the-art pathology foundation models while requiring significantly fewer parameters and less pretraining data.
License
The model is licensed under EXAONEPath AI Model License Agreement 1.0 - NC
Citation
If you find EXAONE Path 2.5 useful in your research, please cite:
@misc{yun2025exaonepath25,
title = {EXAONE Path 2.5: Pathology Foundation Model with Multi-Omics Alignment},
author = {Yun, Juseung and Yu, Sunwoo and Ha, Sumin and Kim, Jonghyun and Lee, Janghyeon and Jang, Jongseong and Lee, Soonyoung},
year = {2025},
eprint = {2512.14019},
archivePrefix= {arXiv},
primaryClass = {cs.LG},
url = {https://arxiv.org/abs/2512.14019}
}
Contact
LG AI Research Technical Support: [email protected]